-
World Health Organization. https://www.who.int/news/item/05-05-2022-14.9-million-excess-deaths-were-associated-with-the-covid-19-pandemic-in-2020-and-2021WHO (2022).
-
Gorbalenya, A. E. et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544 (2020).
-
Peacock, T. P., Penrice-Randal, R., Hiscox, J. A. & Barclay, W. S. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J. Gen. Virol. https://doi.org/10.1099/jgv.0.001584 (2021).
-
Lu, L. et al. Adaptation, spread and transmission of SARS-CoV-2 in farmed minks and associated humans in the Netherlands. Nat. Commun. 12, 6802 (2021).
-
Marques, A. D. et al. Multiple introductions of SARS-CoV-2 Alpha and Delta variants into white-tailed deer in Pennsylvania. mBio 13, e02101-22 (2022).
-
Hale, V. L. et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 602, 481–486 (2022).
-
Bashor, L. et al. SARS-CoV-2 evolution in animals suggests mechanisms for rapid variant selection. Proc. Natl Acad. Sci. USA 118, e2105253118 (2021).
-
MacLean, O. A., Orton, R. J., Singer, J. B. & Robertson, D. L. No evidence for distinct types in the evolution of SARS-CoV-2. Virus Evolut. https://doi.org/10.1093/ve/veaa034 (2020).
-
Volz, E. et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184, 64–75.e11 (2021).
-
O’Toole, Á., Pybus, O. G., Abram, M. E., Kelly, E. J. & Rambaut, A. Pango lineage designation and assignment using SARS-CoV-2 spike gene nucleotide sequences. BMC Genom. 23, 121–121 (2022).
-
Harvey, W. T. et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424 (2021).
-
Woolhouse, M. E., Taylor, L. H. & Haydon, D. T. Population biology of multihost pathogens. Science 292, 1109–1112 (2001).
-
Parrish, C. R. et al. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72, 457–470 (2008).
-
MacLean, O. A. et al. Natural selection in the evolution of SARS-CoV-2 in bats created a generalist virus and highly capable human pathogen. PLoS Biol. 19, e3001115 (2021).
-
Conceicao, C. et al. The SARS-CoV-2 spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol. 18, e3001016 (2020).
-
Delaune, D. et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat. Commun. 12, 6563 (2021).
-
Lin, X. D. et al. Extensive diversity of coronaviruses in bats from China. Virology 507, 1–10 (2017).
-
Starr, T. N. et al. ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Nature 603, 913–918 (2022).
-
Peacock, T. P. et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 6, 899–909 (2021).
-
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020).
-
Zhu, Y. et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 12, 961 (2021).
-
Peacock, T. P. et al. The SARS-CoV-2 variants associated with infections in India, B.1.617, show enhanced spike cleavage by furin. Preprint at bioRxiv https://doi.org/10.1101/2021.05.28.446163 (2021).
-
Mlcochova, P. et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599, 114–119 (2021).
-
Lubinski, B. et al. Spike protein cleavage-activation in the context of the SARS-CoV-2 P681R mutation: an analysis from its first appearance in lineage A.23.1 identified in Uganda. Microbiol. Spectr. 10, e0151422 (2022).
-
Brown, J. C. et al. Increased transmission of SARS-CoV-2 lineage B.1.1.7 (VOC 2020212/01) is not accounted for by a replicative advantage in primary airway cells or antibody escape. Preprint at bioRxiv https://doi.org/10.1101/2021.02.24.432576 (2021).
-
Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310.e20 (2020).
-
Liu, Y. et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 602, 294–299 (2022).
-
Campbell, F. et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Eurosurveillance 26, 2100509 (2021).
-
Davies, N. G. et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372, eabg3055 (2021).
-
Liu, Y. et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 39, 110829 (2022).
-
Willett, B. J. et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. https://doi.org/10.1038/s41564-022-01143-7 (2022).
-
Peacock, T. P. et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. Preprint at bioRxiv https://doi.org/10.1101/2021.12.31.474653 (2022).
-
Cao, Y. et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663 (2022).
-
Zhou, J. et al. Omicron breakthrough infections in vaccinated or previously infected hamsters. Preprint at bioRxiv https://doi.org/10.1101/2022.05.20.492779 (2022).
-
Thomson, E. C. et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187.e20 (2021).
-
Hsieh, C.-L. et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020).
-
Garcia-Beltran, W. F. et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184, 2372–2383.e9 (2021).
-
Newman, J. et al. Neutralizing antibody activity against 21 SARS-CoV-2 variants in older adults vaccinated with BNT162b2. Nat. Microbiol. 7, 1180–1188 (2022).
-
Davis, C. et al. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathog. 17, e1010022 (2021).
-
Lopez Bernal, J. et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N. Engl. J. Med. 385, 585–594 (2021).
-
Andrews, N. et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. https://doi.org/10.1056/NEJMoa2119451 (2022).
-
Abu-Raddad, L. J., Chemaitelly, H. & Butt, A. A. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N. Engl. J. Med. 385, 187–189 (2021).
-
Tegally, H. et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. https://doi.org/10.1038/s41591-022-01911-2 (2022).
-
Viana, R. et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature https://doi.org/10.1038/s41586-022-04411-y (2022).
-
Cele, S. et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 602, 654–656 (2022).
-
Willett, B. J. et al. Distinct antigenic properties of the SARS-CoV-2 Omicron lineages BA.4 and BA.5. Preprint at bioRxiv https://doi.org/10.1101/2022.05.25.493397 (2022).
-
Tuekprakhon, A. et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell https://doi.org/10.1016/j.cell.2022.06.005 (2022).
-
Cao, Y. et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature https://doi.org/10.1038/s41586-022-04980-y (2022).
-
Khan, K. et al. Omicron BA.4/BA.5 escape neutralizing immunity elicited by BA.1 infection. Nat. Commun. 13, 4686 (2022).
-
Meng, B. et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature https://doi.org/10.1038/s41586-022-04474-x (2022).
-
Simon-Loriere, E. & Schwartz, O. Towards SARS-CoV-2 serotypes? Nat. Rev. Microbiol. 20, 187–188 (2022).
-
van der Straten, K. et al. Antigenic cartography using sera from sequence-confirmed SARS-CoV-2 variants of concern infections reveals antigenic divergence of Omicron. Immunity https://doi.org/10.1016/j.immuni.2022.07.018 (2022).
-
Accorsi, E. K. et al. Association between 3 doses of mRNA COVID-19 vaccine and symptomatic infection caused by the SARS-CoV-2 Omicron and Delta variants. JAMA 327, 639–651 (2022).
-
Kherabi, Y., Launay, O. & Luong Nguyen, L. B. COVID-19 vaccines against Omicron variant: real-world data on effectiveness. Viruses 14, 2086 (2022).
-
Kirsebom, F. C. M. et al. COVID-19 vaccine effectiveness against the omicron (BA.2) variant in England. Lancet Infect. Dis. https://doi.org/10.1016/S1473-3099(22)00309-7 (2022).
-
Ferdinands, J. M. et al. Waning 2-dose and 3-dose effectiveness of mRNA vaccines against COVID-19-associated emergency department and urgent care encounters and hospitalizations among adults during periods of Delta and Omicron variant predominance – VISION Network, 10 states, August 2021-January 2022. MMWR Morb. Mortal. Wkly Rep. 71, 255–263 (2022).
-
Collie, S., Champion, J., Moultrie, H., Bekker, L.-G. & Gray, G. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa. N. Engl. J. Med. 386, 494–496 (2021).
-
Higdon, M. M. et al. Duration of effectiveness of vaccination against COVID-19 caused by the omicron variant. Lancet Infect. Dis. 22, 1114–1116 (2022).
-
Fang, Z. et al. Omicron-specific mRNA vaccination alone and as a heterologous booster against SARS-CoV-2. Nat. Commun. 13, 3250 (2022).
-
Alu, A. et al. Intranasal COVID-19 vaccines: from bench to bed. eBioMedicine 76, 103841 (2022).
-
Dolgin, E. Pan-coronavirus vaccine pipeline takes form. Nat. Rev. Drug Discov. 21, 324–326 (2022).
-
Eguia, R. T. et al. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog. 17, e1009453 (2021).
-
Edridge, A. W. D. et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 26, 1691–1693 (2020).
-
Hoffmann, M., Kleine-Weber, H. & Pöhlmann, S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78, 779–784.e5 (2020).
-
Johnson, B. A. et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591, 293–299 (2021).
-
Misumi, Y. et al. Functional expression of furin demonstrating its intracellular localization and endoprotease activity for processing of proalbumin and complement pro-C3. J. Biol. Chem. 266, 16954–16959 (1991).
-
Hill, V. et al. The origins and molecular evolution of SARS-CoV-2 lineage B.1.1.7 in the UK. Virus Evol. https://doi.org/10.1093/ve/veac080 (2022).
-
Laiton-Donato, K. et al. Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Infect. Genet. Evol. 95, 105038 (2021).
-
Saito, A. et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 602, 300–306 (2022).
-
Lubinski, B., Jaimes, J. A. & Whittaker, G. R. Intrinsic furin-mediated cleavability of the spike S1/S2 site from SARS-CoV-2 variant B.1.1.529 (Omicron). Preprint at bioRxiv https://doi.org/10.1101/2022.04.20.488969 (2022).
-
Lubinski, B. et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 25, 103589 (2022).
-
Zhang, L. et al. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc. Natl Acad. Sci. USA 118, e2109905118 (2021).
-
Sanda, M., Morrison, L. & Goldman, R. N- and O-glycosylation of the SARS-CoV-2 spike protein. Anal. Chem. 93, 2003–2009 (2021).
-
Gonzalez-Rodriguez, E. et al. O-linked sialoglycans modulate the proteolysis of SARS-CoV-2 spike and contribute to the mutational trajectory in variants of concern. Preprint at bioRxiv https://doi.org/10.1101/2022.09.15.508093 (2022).
-
Suzuki, R. et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature https://doi.org/10.1038/s41586-022-04462-1 (2022).
-
Mesner, D. et al. SARS-CoV-2 Spike evolution influences GBP and IFITM sensitivity. Preprint at bioRxiv https://doi.org/10.1101/2022.03.07.481785 (2022).
-
Reuschl, A.-K. et al. Enhanced innate immune suppression by SARS-CoV-2 Omicron subvariants BA.4 and BA.5. Preprint at bioRxiv https://doi.org/10.1101/2022.07.12.499603 (2022).
-
Hui, K. P. Y. et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature https://doi.org/10.1038/s41586-022-04479-6 (2022).
-
Aggarwal, A. et al. Platform for isolation and characterization of SARS-CoV-2 variants enables rapid characterization of Omicron in Australia. Nat. Microbiol. 7, 896–908 (2022).
-
Barut, T. et al. The spike gene is a major determinant for the SARS-CoV-2 Omicron-BA.1 phenotype. Preprint at bioRxiv https://doi.org/10.1101/2022.04.28.489537 (2022).
-
Bentley, E. G. et al. SARS-CoV-2 Omicron-B.1.1.529 Variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19. Preprint at bioRxiv https://doi.org/10.1101/2021.12.26.474085 (2021).
-
Kimura, I. et al. SARS-CoV-2 spike S375F mutation characterizes the Omicron BA.1 variant. Preprint at bioRxiv https://doi.org/10.1101/2022.04.03.486864 (2022).
-
Yamamoto, M. et al. SARS-CoV-2 Omicron spike H655Y mutation is responsible for enhancement of the endosomal entry pathway and reduction of cell surface entry pathways. Preprint at bioRxiv https://doi.org/10.1101/2022.03.21.485084 (2022).
-
Kimura, I. et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants including BA.4 and BA.5. Cell https://doi.org/10.1016/j.cell.2022.09.018 (2022).
-
Allen, H. et al. Comparative transmission of SARS-CoV-2 Omicron (B.1.1.529) and Delta (B.1.617.2) variants and the impact of vaccination: national cohort study, England. Preprint at medRxiv https://doi.org/10.1101/2022.02.15.22271001 (2022).
-
Lyngse, F. P. et al. Household transmission of the SARS-CoV-2 Omicron variant in Denmark. Nat. Commun. 13, 5573 (2022).
-
Yu, S. et al. SARS-CoV-2 spike engagement of ACE2 primes S2′ site cleavage and fusion initiation. Proc. Natl Acad. Sci. USA 119, e2111199119 (2022).
-
Millet, J. K. & Whittaker, G. R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl Acad. Sci. USA 111, 15214–15219 (2014).
-
Belouzard, S., Chu, V. C. & Whittaker, G. R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl Acad. Sci. USA 106, 5871–5876 (2009).
-
Benton, D. J. et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 588, 327–330 (2020).
-
Bestle, D. et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 3, e202000786 (2020).
-
Qing, E. et al. Inter-domain communication in SARS-CoV-2 spike proteins controls protease-triggered cell entry. Cell Rep. 39, 110786 (2022).
-
Meng, B. et al. SARS-CoV-2 spike N-terminal domain modulates TMPRSS2-dependent viral entry and fusogenicity. Cell Rep. 40, 111220 (2022).
-
Cantoni, D. et al. Evolutionary remodelling of N-terminal domain loops fine-tunes SARS-CoV-2 spike. EMBO Rep. 23, e54322 (2022).
-
Syed, A. M. et al. Rapid assessment of SARS-CoV-2-evolved variants using virus-like particles. Science 374, 1626–1632 (2021).
-
Wu, H. et al. Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2. Cell Host Microbe 29, 1788–1801.e6 (2021).
-
Johnson, B. A. et al. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLoS Pathog. 18, e1010627 (2022).
-
Mears, H. V. et al. Emergence of new subgenomic mRNAs in SARS-CoV-2. Preprint at bioRxiv https://doi.org/10.1101/2022.04.20.488895 (2022).
-
Leary, S. et al. Generation of a novel SARS-CoV-2 sub-genomic RNA due to the R203K/G204R variant in nucleocapsid: homologous recombination has potential to change SARS-CoV-2 at both protein and RNA level. Pathog. Immun. 6, 27–49 (2021).
-
Syed, A. M. et al. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. Proc. Natl Acad. Sci. USA 119, e2200592119 (2022).
-
Schoeman, D. & Fielding, B. C. Coronavirus envelope protein: current knowledge. Virol. J. 16, 69 (2019).
-
Xia, B. et al. Why SARS-CoV-2 Omicron variant is milder? A single high-frequency mutation of structural envelope protein matters. Preprint at bioRxiv https://doi.org/10.1101/2022.02.01.478647 (2022).
-
Ricciardi, S. et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature https://doi.org/10.1038/s41586-022-04835-6 (2022).
-
Peng, Y. et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 21, 1336–1345 (2020).
-
Moss, P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 23, 186–193 (2022).
-
Feng, S. et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 27, 2032–2040 (2021).
-
Tan, A. T. et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 34, 108728 (2021).
-
Bange, E. M. et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 27, 1280–1289 (2021).
-
Oberhardt, V. et al. Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature 597, 268–273 (2021).
-
Tarke, A. et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2, 100204 (2021).
-
Agerer, B. et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8+ T cell responses. Sci. Immunol. 6, eabg6461 (2021).
-
Goonetilleke, N. et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206, 1253–1272 (2009).
-
Wilkinson, S. A. J. et al. Recurrent SARS-CoV-2 mutations in immunodeficient patients. Virus Evol. https://doi.org/10.1093/ve/veac050 (2022).
-
Stanevich, O. et al. SARS-CoV-2 escape from cytotoxic T cells during long-term COVID-19. Preprint at Res. Sq. https://doi.org/10.21203/rs.3.rs-750741/v1 (2021).
-
de Silva, T. I. et al. The impact of viral mutations on recognition by SARS-CoV-2 specific T cells. iScience 24, 103353 (2021).
-
Motozono, C. et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 29, 1124–1136.e11 (2021).
-
Dolton, G. et al. Emergence of immune escape at dominant SARS-CoV-2 killer T cell epitope. Cell 185, 2936–2951.e19 (2022).
-
Riou, C. et al. Escape from recognition of SARS-CoV-2 variant spike epitopes but overall preservation of T cell immunity. Sci. Transl Med. 14, eabj6824 (2022).
-
Reynolds, C. J. et al. Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science 372, 1418–1423 (2021).
-
Tarke, A. et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 185, 847–859.e11 (2022).
-
UKHSA. COVID-19 Vaccine Surveillance Report (UKHSA, 2022).
-
Naranbhai, V. et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell 185, 1041–1051.e6 (2022).
-
Wellington, D. et al. SARS-CoV-2 mutations affect proteasome processing to alter CD8+ T cell responses. Preprint at bioRxiv https://doi.org/10.1101/2022.04.08.487623 (2022).
-
Ladell, K. et al. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity 38, 425–436 (2013).
-
Zhang, Y. et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-I. Proc. Natl Acad. Sci. USA 118, e2024202118 (2021).
-
Moriyama, M., Lucas, C., Monteiro, V. S., Yale SARS-CoV-2 Genomic Surveillance Initiative & Iwasaki, A. SARS-CoV-2 Omicron subvariants evolved to promote further escape from MHC-I recognition. Preprint at bioRxiv https://doi.org/10.1101/2022.05.04.490614 (2022).
-
Yoo, J.-S. et al. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 12, 6602 (2021).
-
Arshad, N. et al. SARS-CoV-2 accessory proteins ORF7a and ORF3a use distinct mechanisms to downregulate MHC-I surface expression. Preprint at bioRxiv https://doi.org/10.1101/2022.05.17.492198 (2022).
-
Zhang, F. et al. Inhibition of major histocompatibility complex-I antigen presentation by sarbecovirus ORF7a proteins. Preprint at bioRxiv https://doi.org/10.1101/2022.05.25.493467 (2022).
-
Stewart, H. et al. Tetherin antagonism by SARS-CoV-2 enhances virus release: multiple mechanisms including ORF3a-mediated defective retrograde traffic. Preprint at bioRxiv https://doi.org/10.1101/2021.01.06.425396 (2021).
-
Rambaut, A. et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological.org https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (2020).
-
Mandl, J. N. et al. Reservoir host immune responses to emerging zoonotic viruses. Cell 160, 20–35 (2015).
-
Thorne, L. G. et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 602, 487–495 (2022).
-
Lista, M. J. et al. The P681H mutation in the spike glycoprotein confers type I interferon resistance in the SARS-CoV-2 alpha (B.1.1.7) variant. Preprint at bioRxiv https://doi.org/10.1101/2021.11.09.467693 (2021).
-
Guo, K. et al. Interferon resistance of emerging SARS-CoV-2 variants. Proc. Natl Acad. Sci. USA 119, e2203760119 (2022).
-
Bouhaddoum, M. et al. Global landscape of the host response to SARS-CoV-2 variants reveals viral evolutionary trajectories. Preprint at bioRxiv https://doi.org/10.1101/2022.10.19.512927 (2022).
-
Hall, R. et al. SARS-CoV-2 ORF6 disrupts innate immune signalling by inhibiting cellular mRNA export. PLoS Pathog. 18, e1010349 (2022).
-
Jiang, H.-w et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 17, 998–1000 (2020).
-
Beyer, D. K. & Forero, A. Mechanisms of antiviral immune evasion of SARS-CoV-2. J. Mol. Biol. 434, 167265 (2022).
-
Xu, D., Biswal, M., Neal, A. & Hai, R. Devil’s tools: SARS-CoV-2 antagonists against innate immunity. Curr. Res. Virol. Sci. 2, 100013 (2021).
-
Shalamova, L. et al. Omicron variant of SARS-CoV-2 exhibits an increased resilience to the antiviral type I interferon response. PNAS Nexus https://doi.org/10.1093/pnasnexus/pgac067 (2022).
-
Winstone, H. et al. The polybasic cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J. Virol. https://doi.org/10.1128/jvi.02422-20 (2021).
-
Zhao, X. et al. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl Acad. Sci. USA 111, 6756–6761 (2014).
-
Prelli Bozzo, C. et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 12, 4584 (2021).
-
Nchioua, R. et al. SARS-CoV-2 variants of concern hijack IFITM2 for efficient replication in human lung cells. J. Virol. 96, e00594-22 (2022).
-
Diamond, M. S. & Farzan, M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 13, 46–57 (2013).
-
Li, K. et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 9, e1003124 (2013).
-
Klein, S. et al. IFITM3 blocks viral entry by sorting lipids and stabilizing hemifusion. Preprint at bioRxiv https://doi.org/10.1101/2022.09.01.506185 (2022).
-
Sandler, N. G. & Douek, D. C. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat. Rev. Microbiol. 10, 655–666 (2012).
-
Wang, Z. et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature 595, 426–431 (2021).
-
Levin, E. G. et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N. Engl. J. Med. 385, e84 (2021).
-
Paton, R. S., Overton, C. E. & Ward, T. The rapid replacement of the Delta variant by Omicron (B.1.1.529) in England. Sci. Transl Med. 14, eabo5395 (2022).
-
Earnest, R. et al. Comparative transmissibility of SARS-CoV-2 variants Delta and Alpha in New England, USA. Cell Rep. Med. 3, 100583 (2022).
-
Tang, P. et al. BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 Delta variant in Qatar. Nat. Med. 27, 2136–2143 (2021).
-
Bruxvoort, K. J. et al. Effectiveness of mRNA-1273 against delta, mu, and other emerging variants of SARS-CoV-2: test negative case-control study. BMJ 375, e068848 (2021).
-
Lyngse, F. P. et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. Preprint at medRxiv https://doi.org/10.1101/2022.01.28.22270044 (2022).
-
Cao, Y. et al. Imprinted SARS-CoV-2 humoral immunity induces converging Omicron RBD evolution. Preprint at bioRxiv https://doi.org/10.1101/2022.09.15.507787 (2022).
-
Sheward, D. J. et al. Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Preprint at bioRxiv https://doi.org/10.1101/2022.09.16.508299 (2022).
-
Sheward, D. J. et al. Evasion of neutralising antibodies by omicron sublineage BA.2.75. Lancet Infect. Dis. https://doi.org/10.1016/S1473-3099(22)00524-2 (2022).
-
Wang, Q. et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 608, 603–608 (2022).
-
Anderson, R. M. & May, R. M. Coevolution of hosts and parasites. Parasitology 85, 411–426 (1982).
-
Alizon, S., Hurford, A., Mideo, N. & Van Baalen, M. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 22, 245–259 (2009).
-
Sasaki, A., Lion, S. & Boots, M. Antigenic escape selects for the evolution of higher pathogen transmission and virulence. Nat. Ecol. Evol. 6, 51–62 (2022).
-
Pascall, D. J. et al. Inconsistent directions of change in case severity across successive SARS-CoV-2 variant waves suggests an unpredictable future. Preprint at medRxiv https://doi.org/10.1101/2022.03.24.22272915 (2022).
-
Wolter, N. et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet 399, 437–446 (2022).
-
Bager, P. et al. Risk of hospitalisation associated with infection with SARS-CoV-2 omicron variant versus delta variant in Denmark: an observational cohort study. Lancet Infect. Dis. https://doi.org/10.1016/s1473-3099(22)00154-2 (2022).
-
Nyberg, T. et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet https://doi.org/10.1016/S0140-6736(22)00462-7 (2022).
-
Lewnard, J. A. et al. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in southern California. Nat. Med. https://doi.org/10.1038/s41591-022-01887-z (2022).
-
Lamers, M. M. & Haagmans, B. L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 20, 270–284 (2022).
-
Dong, W. et al. The K18-human ACE2 transgenic mouse model recapitulates non-severe and severe COVID-19 in response to an infectious dose of the SARS-CoV-2 virus. J. Virol. 96, e0096421 (2022).
-
Thakur, N. et al. SARS-CoV-2 variants of concern alpha, beta, gamma and delta have extended ACE2 receptor host ranges. J. Gen. Virol. https://doi.org/10.1099/jgv.0.001735 (2022).
-
Menachery, V. D., Gralinski, L. E., Baric, R. S. & Ferris, M. T. New metrics for evaluating viral respiratory pathogenesis. PLoS ONE 10, e0131451 (2015).
-
Uraki, R. et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA.2. Nature https://doi.org/10.1038/s41586-022-04856-1 (2022).
-
Halfmann, P. J. et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603, 687–692 (2022).
-
Mefsin, Y. et al. Epidemiology of infections with SARS-CoV-2 Omicron BA.2 variant in Hong Kong, January-March 2022. Preprint at medRxiv https://doi.org/10.1101/2022.04.07.22273595 (2022).
-
Shuai, H. et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603, 693–699 (2022).
-
van Doremalen, N. et al. SARS-CoV-2 Omicron BA.1 and BA.2 are attenuated in rhesus macaques as compared to Delta. Preprint at bioRxiv https://doi.org/10.1101/2022.08.01.502390 (2022).
-
Su, W. et al. Omicron BA.1 and BA.2 sub-lineages show reduced pathogenicity and transmission potential than the early SARS-CoV-2 D614G variant in Syrian hamsters. J. Infect. Dis. https://doi.org/10.1093/infdis/jiac276 (2022).
-
Hu, B., Guo, H., Zhou, P. & Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 19, 141–154 (2021).
-
Niemi, M. E. K., Daly, M. J. & Ganna, A. The human genetic epidemiology of COVID-19. Nat. Rev. Genet. https://doi.org/10.1038/s41576-022-00478-5 (2022).
-
Whitaker, M. et al. Variant-specific symptoms of COVID-19 among 1,542,510 people in England. Preprint at medRxiv https://doi.org/10.1101/2022.05.21.22275368 (2022).
-
Gonzalez-Reiche, A. S. et al. SARS-CoV-2 variants in the making: Sequential intrahost evolution and forward transmissions in the context of persistent infections. Preprint at medRxiv https://doi.org/10.1101/2022.05.25.22275533 (2022).
-
Ghafari, M., Liu, Q., Dhillon, A., Katzourakis, A. & Weissman, D. B. Investigating the evolutionary origins of the first three SARS-CoV-2 variants of concern. Front. Virol. https://doi.org/10.3389/fviro.2022.942555 (2022).
-
Harari, S. et al. Drivers of adaptive evolution during chronic SARS-CoV-2 infections. Nat. Med. https://doi.org/10.1038/s41591-022-01882-4 (2022).
-
Roemer, C. Variant report 2022-08-31. https://github.com/neherlab/SARS-CoV-2_variant-reports/tree/main/reports (2022).
-
Pickering, B. et al. Highly divergent white-tailed deer SARS-CoV-2 with potential deer-to-human transmission. Preprint at bioRxiv https://doi.org/10.1101/2022.02.22.481551 (2022).
-
McCallum, M. et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 184, 2332–2347.e16 (2021).
-
Barnes, C. O. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020).
-
Greaney, A. J. et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476.e6 (2021).
-
Lan, J. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020).
-
Gao, Y. et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 28, 472–476 (2022).
-
Wright, D. W. et al. Tracking SARS-CoV-2 mutations and variants through the COG-UK-Mutation Explorer. Virus Evol. https://doi.org/10.1093/ve/veac023 (2022).
-
Faria, N. R. et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 372, 815–821 (2021).
-
Tegally, H. et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592, 438–443 (2021).
-
Dhar, M. S. et al. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Science 374, 995–999 (2021).
-
McCrone, J. T. et al. Context-specific emergence and growth of the SARS-CoV-2 Delta variant. Nature https://doi.org/10.1038/s41586-022-05200-3 (2022).
-
UKHSA. SARS-CoV-2 Variants of Concern and Variants Under Investigation in England: Technical Briefing 25 (UKHSA, 2021).
-
Mallapaty, S. Where did Omicron come from? Three key theories. Nature 602, 26–28 (2022).
-
Choi, B. et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 383, 2291–2293 (2020).
-
Chaguza, C. et al. Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection. Preprint at medRxiv https://doi.org/10.1101/2022.06.29.22276868 (2022).
-
Kemp, S. A. et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 592, 277–282 (2021).
-
Jackson, B. et al. Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell 184, 5179–5188.e8 (2021).
-
Sekizuka, T. et al. Genome recombination between Delta and Alpha variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Jpn J. Infect. Dis. https://doi.org/10.7883/yoken.JJID.2021.844 (2022).
-
Simon-Loriere, E. et al. Rapid characterization of a Delta-Omicron SARS-CoV-2 recombinant detected in Europe. Preprint at Res. Sq. https://doi.org/10.21203/rs.3.rs-1502293/v1 (2022).
-
UKHSA. SARS-CoV-2 Variants of Concern and Variants Under Investigation in England: Technical Briefing 40 (UKHSA, 2022).
-
Network for Surveillance-South Africa (NGS-SA). SARS-CoV-2 Sequencing Update 15 July 2022 (NGS-SA, 2022).
-
Cao, Y. et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Preprint at bioRxiv https://doi.org/10.1101/2022.09.15.507787 (2022).
-
Lytras, S. et al. Exploring the natural origins of SARS-CoV-2 in the light of recombination. Genome Biol. Evol. https://doi.org/10.1093/gbe/evac018 (2022).
-
Demetrius, L. & Ziehe, M. Darwinian fitness. Theor. Popul. Biol. 72, 323–345 (2007).
-
Domingo, E. & Holland, J. J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51, 151–178 (1997).
-
Wargo, A. R. & Kurath, G. Viral fitness: definitions, measurement, and current insights. Curr. Opin. Virol. 2, 538–545 (2012).
Page 2
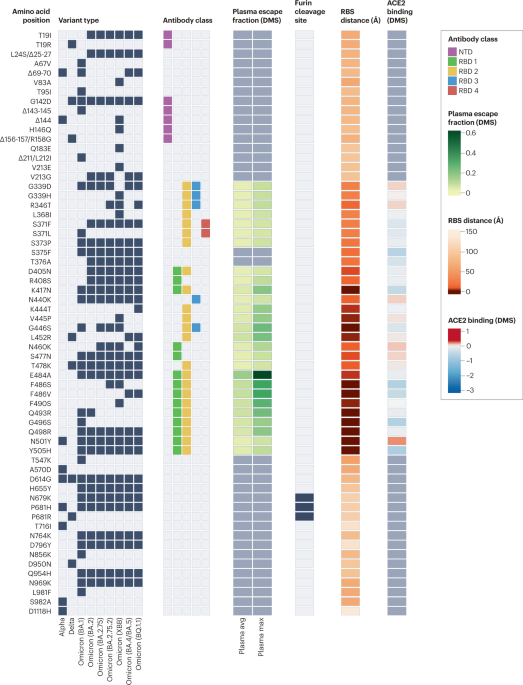
Black boxes denote the presence of each mutation in the variant of concern. Epitope residues are coloured to indicate the amino-terminal domain (NTD) supersite187 or the receptor-binding domain (RBD) class188. For RBD residues, the results of deep mutational scanning (DMS) studies show the escape fraction (that is, a quantitative measure of the extent to which a mutation reduced polyclonal antibody binding) for each mutant averaged across plasma (‘plasma avg’) and for the most sensitive plasma (‘plasma max’)189, illustrating consistency or variation in the effect of a mutation depending on differences in the antibody repertoire of individuals. Mutations in the furin cleavage site are highlighted. Orange shading indicates the distance to angiotensin-converting enzyme 2 (ACE2)-contacting residues that form the receptor-binding site (RBS). Note that the RBS is defined as residues with an atom <4 Å from an ACE2 atom in the structure of the RBD bound to ACE2 (RCSB Protein Data Bank ID 6M0J190). Finally, ACE2-binding scores representing the binding constant (Δlog10 KD) relative to the wild-type reference amino acid from DMS experiments are shown in shades of red or blue26.

